Welcome to Our Bioinformatics Lab!
Bioinformatics Lab
Welcome to Bioinformatics Lab
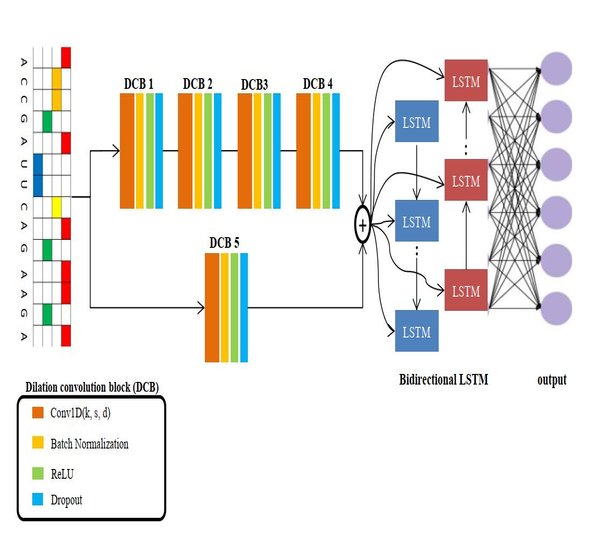
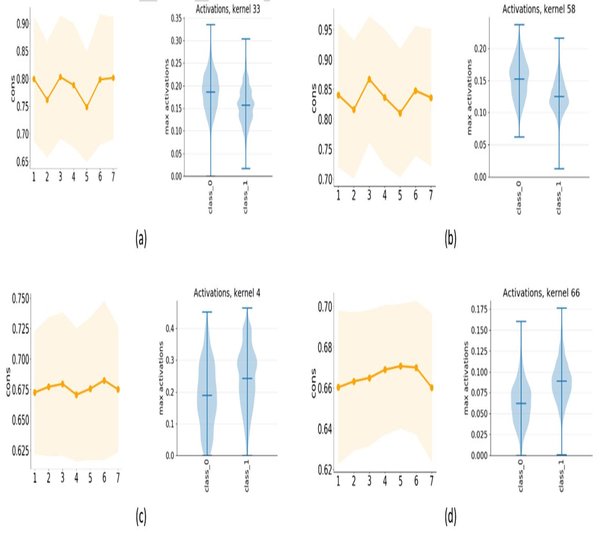
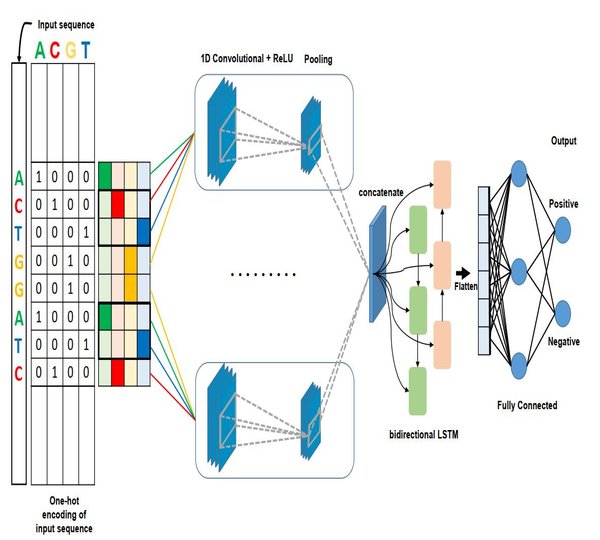
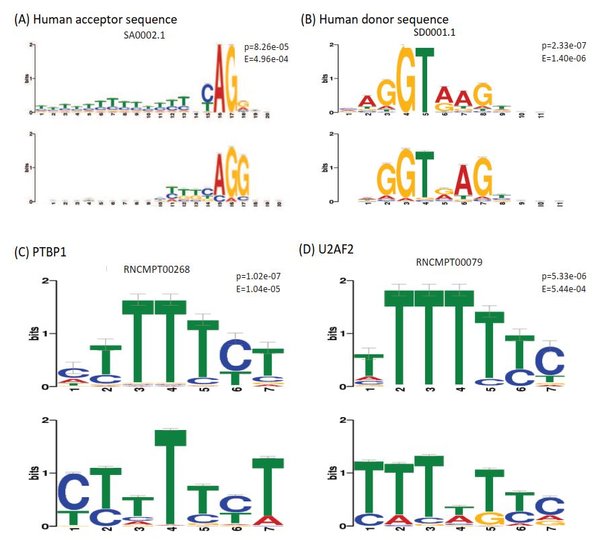
Our lab is part of the Electronics Engineering Faculty in JeonBuk National University and we have put a lot of effort on preparing not only excellent employees of domestic IT industry but human resource development with international competitiveness and leadership. Our lab disciplines is to build computational systems for Transciption Factor Binding Site, Alternative Splicing, Branch Point Selection, etc.
Our lab goal is training industry's most wanted human resource with specialized knowledge and practical skills, excellent researcher with creativity, globally competitive advanced human resource. Many students involved in our lab in a variety of programs which are provided by our university and faculty, such as Industry Internship program, Professor Laboratory Practicum program etc. Graduates are contributing IT industry development greatly from wide range of sectors including enterprises, research institutes and universities.
Read More